📥 Visualizing correlated Gaussian distributions#
A multivariate Gaussian distribution for \(N\) dimensional \(\boldsymbol{x} = \{x_1, \ldots, x_N\}\) with \(\boldsymbol{\mu} = \{\mu_1, \ldots, \mu_N\}\), with positive-definite covariance matrix \(\Sigma\) is
For the one dimensional case, it reduces to the familiar
with \(\Sigma = \sigma_1^2\).
For the bivariate case (two dimensional),
and \(\Sigma\) is positive definite.
Questions to consider:
What does “positive definite” mean and why is this a requirement for the covariance matrix \(\Sigma\)?
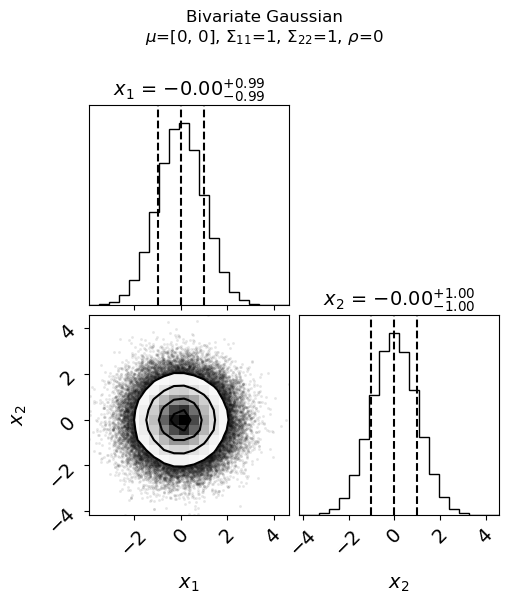
What is plotted in each part of the graph (called a “corner plot”)?
What effect does changing \(\mu_1\) (
mu1) or \(\mu_2\) (mu2) have?What effect does changing \(\sigma_1\) (
var1) or \(\sigma_2\) (var2) have? What if the scales for \(x_1\) and \(x_2\) were the same?What happens if \(\rho_{12}\) (
rho) is equal to \(0\) then \(+0.5\) then \(-0.5\).What happens if \(\rho_{12}\) (
rho) is \(>1\) or \(< -1\)? Explain what goes wrong.So what characterizes independent (uncorrelated) variables versus positively correlated versus negatively correlated?
import numpy as np
import matplotlib.pyplot as plt
import corner
def plot_bivariate_gaussians(mu1, mu2, var1, var2, rho, samples=100000):
"""
Generates and plots samples from a 2D correlated Gaussian distribution
(also known as a bivariate normal distribution).
Args:
mu1 (float): Mean of the first dimension.
mu2 (float): Mean of the second dimension.
var1 (float): Variance of the first dimension.
var2 (float): Variance of the second dimension.
rho (float): Correlation coefficient between the dimensions.
"""
# Create the mean vector and covariance matrix
mean = np.array([mu1, mu2])
cov = np.array([
[var1, rho * np.sqrt(var1 * var2)],
[rho * np.sqrt(var1 * var2), var2]
])
# Generate samples from the multivariate normal distribution
try:
samples = np.random.multivariate_normal(mean, cov, size=samples)
except np.linalg.LinAlgError:
print("Invalid covariance matrix. Ensure `var1` and `var2` are positive.")
return
# Create the corner plot
fig = corner.corner(samples, labels=[r'$x_1$', r'$x_2$'],
quantiles=[0.16, 0.5, 0.84],
show_titles=True,
label_kwargs={"fontsize": 14},
title_kwargs={"fontsize": 14})
# Adjust the fonts for the tick labels
for ax in fig.get_axes():
ax.tick_params(axis='both', labelsize=14)
# Update the title with the current parameters
fig.suptitle(
f"Bivariate Gaussian\n$\mu$=[{mu1}, {mu2}], $\Sigma_{{11}}$={var1}, $\Sigma_{{22}}$={var2}, $\\rho$={rho}",
y=1.1)
plt.show()
mu1 = 0
mu2 = 0
var1 = 1
var2 = 1
rho = 0
plot_bivariate_gaussians(mu1, mu2, var1, var2, rho)