Gaussian Process Regression#
This notebook uses scikit-learn with the workflow:
build an RBF-based kernel (with signal scale and noise term),
fit the GP on a subset (e.g., every 3rd point),
predict mean and uncertainty on a target grid or the full input,
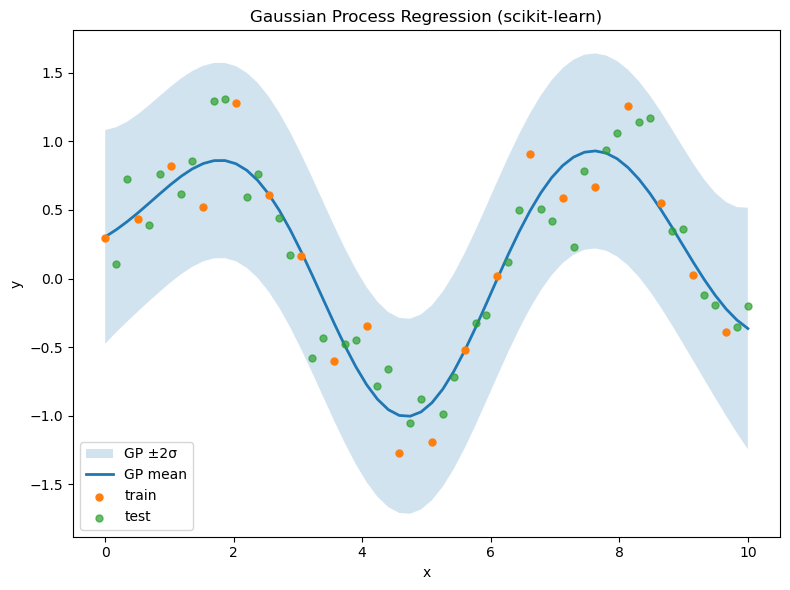
plot mean ±2σ and data,
compute simple validation metrics.
# Imports
import numpy as np
import matplotlib.pyplot as plt
from sklearn.gaussian_process import GaussianProcessRegressor
from sklearn.gaussian_process.kernels import RBF, ConstantKernel as C, WhiteKernel
np.random.seed(1234)
x = np.linspace(0, 10, 60)
f = np.sin(x) + 0.2*np.cos(3*x)
y = f + 0.2*np.random.randn(x.size)
# Ensure shapes (n,1) and (n,)
X = np.asarray(x).reshape(-1, 1)
y = np.asarray(y).reshape(-1)
print('Data shapes -> X:', X.shape, ' y:', y.shape)
Data shapes -> X: (60, 1) y: (60,)
# --- Train/validation split: every 3rd point for train ---
idx = np.arange(X.shape[0])
train_mask = (idx % 3 == 0)
test_mask = ~train_mask
X_train, y_train = X[train_mask], y[train_mask]
X_test, y_test = X[test_mask], y[test_mask]
print('Train size:', X_train.shape[0], ' Test size:', X_test.shape[0])
Train size: 20 Test size: 40
# --- Kernel: signal variance * RBF(length_scale) + white noise ---
kernel = C(1.0, (1e-3, 1e3)) * RBF(length_scale=1.0, length_scale_bounds=(1e-1, 1e3)) + WhiteKernel(noise_level=1e-3, noise_level_bounds=(1e-6, 1e1))
gpr = GaussianProcessRegressor(kernel=kernel, normalize_y=True, random_state=1234, n_restarts_optimizer=9)
print('Initial kernel:', gpr.kernel)
Initial kernel: 1**2 * RBF(length_scale=1) + WhiteKernel(noise_level=0.001)
# --- Fit (replaces GPy: m.optimize(...)) ---
gpr.fit(X_train, y_train)
print('\nOptimized kernel:', gpr.kernel_)
Optimized kernel: 1.03**2 * RBF(length_scale=1.36) + WhiteKernel(noise_level=0.193)
# --- Predict on full X (replaces: yp, vp = m.predict(xp)) ---
y_mean, y_std = gpr.predict(X, return_std=True)
# For test set metrics
from sklearn.metrics import r2_score, mean_absolute_percentage_error
y_pred_test, y_std_test = gpr.predict(X_test, return_std=True)
print('\nValidation:')
print(' R^2 (test):', r2_score(y_test, y_pred_test))
try:
print(' MAPE (test):', mean_absolute_percentage_error(y_test, y_pred_test))
except Exception:
pass
Validation:
R^2 (test): 0.8596309559662871
MAPE (test): 0.4463261384340149
# --- Plot (replaces: m.plot(...)) ---
plt.figure(figsize=(8,6))
# 95% confidence band
plt.fill_between(X.ravel(),
y_mean - 2*y_std,
y_mean + 2*y_std,
alpha=0.2, label='GP ±2σ')
plt.plot(X.ravel(), y_mean, lw=2, label='GP mean')
plt.scatter(X_train.ravel(), y_train, s=25, label='train', zorder=3)
plt.scatter(X_test.ravel(), y_test, s=25, label='test', alpha=0.7, zorder=2)
plt.xlabel('x')
plt.ylabel('y')
plt.title('Gaussian Process Regression (scikit-learn)')
plt.legend()
plt.tight_layout()
plt.show()